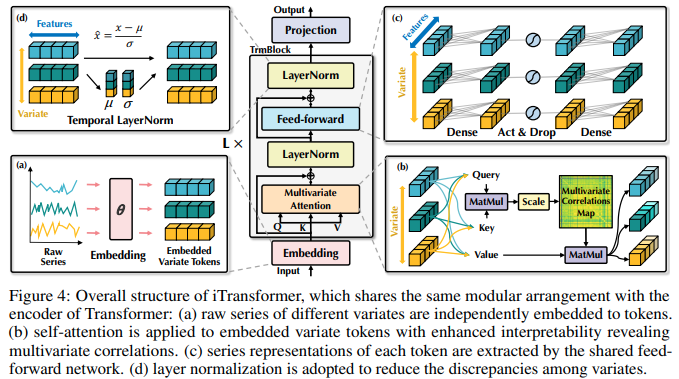
Implementation of iTransformer – SOTA Time Series Forecasting using Attention networks, out of Tsinghua / Ant group
All that remains is tabular data (xgboost still champion here) before one can truly declare “Attention is all you need”
In before Apple gets the authors to change the name.
The official implementation has been released here!
-
StabilityAI and 🤗 Huggingface for the generous sponsorship, as well as my other sponsors, for affording me the independence to open source current artificial intelligence techniques.
-
Greg DeVos for sharing experiments he ran on
iTransformerand some of the improvised variants
$ pip install iTransformer
import torch
from iTransformer import iTransformer
# using solar energy settings
model = iTransformer(
num_variates = 137,
lookback_len = 96, # or the lookback length in the paper
dim = 256, # model dimensions
depth = 6, # depth
heads = 8, # attention heads
dim_head = 64, # head dimension
pred_length = (12, 24, 36, 48), # can be one prediction, or many
num_tokens_per_variate = 1, # experimental setting that projects each variate to more than one token. the idea is that the network can learn to divide up into time tokens for more granular attention across time. thanks to flash attention, you should be able to accommodate long sequence lengths just fine
use_reversible_instance_norm = True # use reversible instance normalization, proposed here https://openreview.net/forum?id=cGDAkQo1C0p . may be redundant given the layernorms within iTransformer (and whatever else attention learns emergently on the first layer, prior to the first layernorm). if i come across some time, i'll gather up all the statistics across variates, project them, and condition the transformer a bit further. that makes more sense
)
time_series = torch.randn(2, 96, 137) # (batch, lookback len, variates)
preds = model(time_series)
# preds -> Dict[int, Tensor[batch, pred_length, variate]]
# -> (12: (2, 12, 137), 24: (2, 24, 137), 36: (2, 36, 137), 48: (2, 48, 137))
For an improvised version that does granular attention across time tokens (as well as the original per-variate tokens), just import iTransformer2D and set the additional num_time_tokens
Update: It works! Thanks goes out to Greg DeVos for running the experiment here!
Update 2: Got an email. Yes you are free to write a paper on this, if the architecture holds up for your problem. I have no skin in the game
import torch
from iTransformer import iTransformer2D
# using solar energy settings
model = iTransformer2D(
num_variates = 137,
num_time_tokens = 16, # number of time tokens (patch size will be (look back length // num_time_tokens))
lookback_len = 96, # the lookback length in the paper
dim = 256, # model dimensions
depth = 6, # depth
heads = 8, # attention heads
dim_head = 64, # head dimension
pred_length = (12, 24, 36, 48), # can be one prediction, or many
use_reversible_instance_norm = True # use reversible instance normalization
)
time_series = torch.randn(2, 96, 137) # (batch, lookback len, variates)
preds = model(time_series)
# preds -> Dict[int, Tensor[batch, pred_length, variate]]
# -> (12: (2, 12, 137), 24: (2, 24, 137), 36: (2, 36, 137), 48: (2, 48, 137))
A iTransformer but also with fourier tokens (FFT of time series is projected into tokens of their own and attended along side with the variate tokens, spliced out at the end)
import torch
from iTransformer import iTransformerFFT
# using solar energy settings
model = iTransformerFFT(
num_variates = 137,
lookback_len = 96, # or the lookback length in the paper
dim = 256, # model dimensions
depth = 6, # depth
heads = 8, # attention heads
dim_head = 64, # head dimension
pred_length = (12, 24, 36, 48), # can be one prediction, or many
num_tokens_per_variate = 1, # experimental setting that projects each variate to more than one token. the idea is that the network can learn to divide up into time tokens for more granular attention across time. thanks to flash attention, you should be able to accommodate long sequence lengths just fine
use_reversible_instance_norm = True # use reversible instance normalization, proposed here https://openreview.net/forum?id=cGDAkQo1C0p . may be redundant given the layernorms within iTransformer (and whatever else attention learns emergently on the first layer, prior to the first layernorm). if i come across some time, i'll gather up all the statistics across variates, project them, and condition the transformer a bit further. that makes more sense
)
time_series = torch.randn(2, 96, 137) # (batch, lookback len, variates)
preds = model(time_series)
# preds -> Dict[int, Tensor[batch, pred_length, variate]]
# -> (12: (2, 12, 137), 24: (2, 24, 137), 36: (2, 36, 137), 48: (2, 48, 137))
- beef up the transformer with latest findings
- improvise a 2d version across both variates and time
- improvise a version that includes fft tokens
- improvise a variant that uses adaptive normalization conditioned on statistics across all variates
@misc{liu2023itransformer,
title = {iTransformer: Inverted Transformers Are Effective for Time Series Forecasting},
author = {Yong Liu and Tengge Hu and Haoran Zhang and Haixu Wu and Shiyu Wang and Lintao Ma and Mingsheng Long},
year = {2023},
eprint = {2310.06625},
archivePrefix = {arXiv},
primaryClass = {cs.LG}
}
@misc{shazeer2020glu,
title = {GLU Variants Improve Transformer},
author = {Noam Shazeer},
year = {2020},
url = {https://arxiv.org/abs/2002.05202}
}
@misc{burtsev2020memory,
title = {Memory Transformer},
author = {Mikhail S. Burtsev and Grigory V. Sapunov},
year = {2020},
eprint = {2006.11527},
archivePrefix = {arXiv},
primaryClass = {cs.CL}
}
@inproceedings{Darcet2023VisionTN,
title = {Vision Transformers Need Registers},
author = {Timoth'ee Darcet and Maxime Oquab and Julien Mairal and Piotr Bojanowski},
year = {2023},
url = {https://api.semanticscholar.org/CorpusID:263134283}
}
@inproceedings{dao2022flashattention,
title = {Flash{A}ttention: Fast and Memory-Efficient Exact Attention with {IO}-Awareness},
author = {Dao, Tri and Fu, Daniel Y. and Ermon, Stefano and Rudra, Atri and R{\'e}, Christopher},
booktitle = {Advances in Neural Information Processing Systems},
year = {2022}
}
@Article{AlphaFold2021,
author = {Jumper, John and Evans, Richard and Pritzel, Alexander and Green, Tim and Figurnov, Michael and Ronneberger, Olaf and Tunyasuvunakool, Kathryn and Bates, Russ and {\v{Z}}{\'\i}dek, Augustin and Potapenko, Anna and Bridgland, Alex and Meyer, Clemens and Kohl, Simon A A and Ballard, Andrew J and Cowie, Andrew and Romera-Paredes, Bernardino and Nikolov, Stanislav and Jain, Rishub and Adler, Jonas and Back, Trevor and Petersen, Stig and Reiman, David and Clancy, Ellen and Zielinski, Michal and Steinegger, Martin and Pacholska, Michalina and Berghammer, Tamas and Bodenstein, Sebastian and Silver, David and Vinyals, Oriol and Senior, Andrew W and Kavukcuoglu, Koray and Kohli, Pushmeet and Hassabis, Demis},
journal = {Nature},
title = {Highly accurate protein structure prediction with {AlphaFold}},
year = {2021},
doi = {10.1038/s41586-021-03819-2},
note = {(Accelerated article preview)},
}
@inproceedings{kim2022reversible,
title = {Reversible Instance Normalization for Accurate Time-Series Forecasting against Distribution Shift},
author = {Taesung Kim and Jinhee Kim and Yunwon Tae and Cheonbok Park and Jang-Ho Choi and Jaegul Choo},
booktitle = {International Conference on Learning Representations},
year = {2022},
url = {https://openreview.net/forum?id=cGDAkQo1C0p}
}
@inproceedings{Katsch2023GateLoopFD,
title = {GateLoop: Fully Data-Controlled Linear Recurrence for Sequence Modeling},
author = {Tobias Katsch},
year = {2023},
url = {https://api.semanticscholar.org/CorpusID:265018962}
}
@article{Zhou2024ValueRL,
title = {Value Residual Learning For Alleviating Attention Concentration In Transformers},
author = {Zhanchao Zhou and Tianyi Wu and Zhiyun Jiang and Zhenzhong Lan},
journal = {ArXiv},
year = {2024},
volume = {abs/2410.17897},
url = {https://api.semanticscholar.org/CorpusID:273532030}
}
@article{Zhu2024HyperConnections,
title = {Hyper-Connections},
author = {Defa Zhu and Hongzhi Huang and Zihao Huang and Yutao Zeng and Yunyao Mao and Banggu Wu and Qiyang Min and Xun Zhou},
journal = {ArXiv},
year = {2024},
volume = {abs/2409.19606},
url = {https://api.semanticscholar.org/CorpusID:272987528}
}